QConWhat?
Why Quantification?
Biological systems are frequently chosen as a target of modelling approaches. Besides measuring the concentration of products and educts of biological reactions and their fluxes, it is often inevitable to gain knowledge of absolute protein abundances to gain a comprehensive understanding. For this, numerous approaches have been proposed, ranging from immuno-plotting and radioactive-plotting to ultra-sensitive high-throughput measurements using mass spectrometry (ms-based proteomics). But even within the field of ms-based proteomics the tastes differ and many absolute quantification methods were proposed in recent years. One method shown to be very practical was the QConCAT technology (even so there is still a significant lack of computational tools!).
Why QConCATs?
To facilitate a certain throughput (and due to lots of other reasons) proteins are not measured directly in so called "shotgun-ms" scenarious but indirectly. This is done by digesting the protein with a known protease, yielding predictable peptides. Sadly, these peptides can often not directly be correlated with a proteins abundance because they can differ in their likelihood to be ionized due do different physico-chemical properties. One way out of this dilemma is the design of QConCAT proteins.
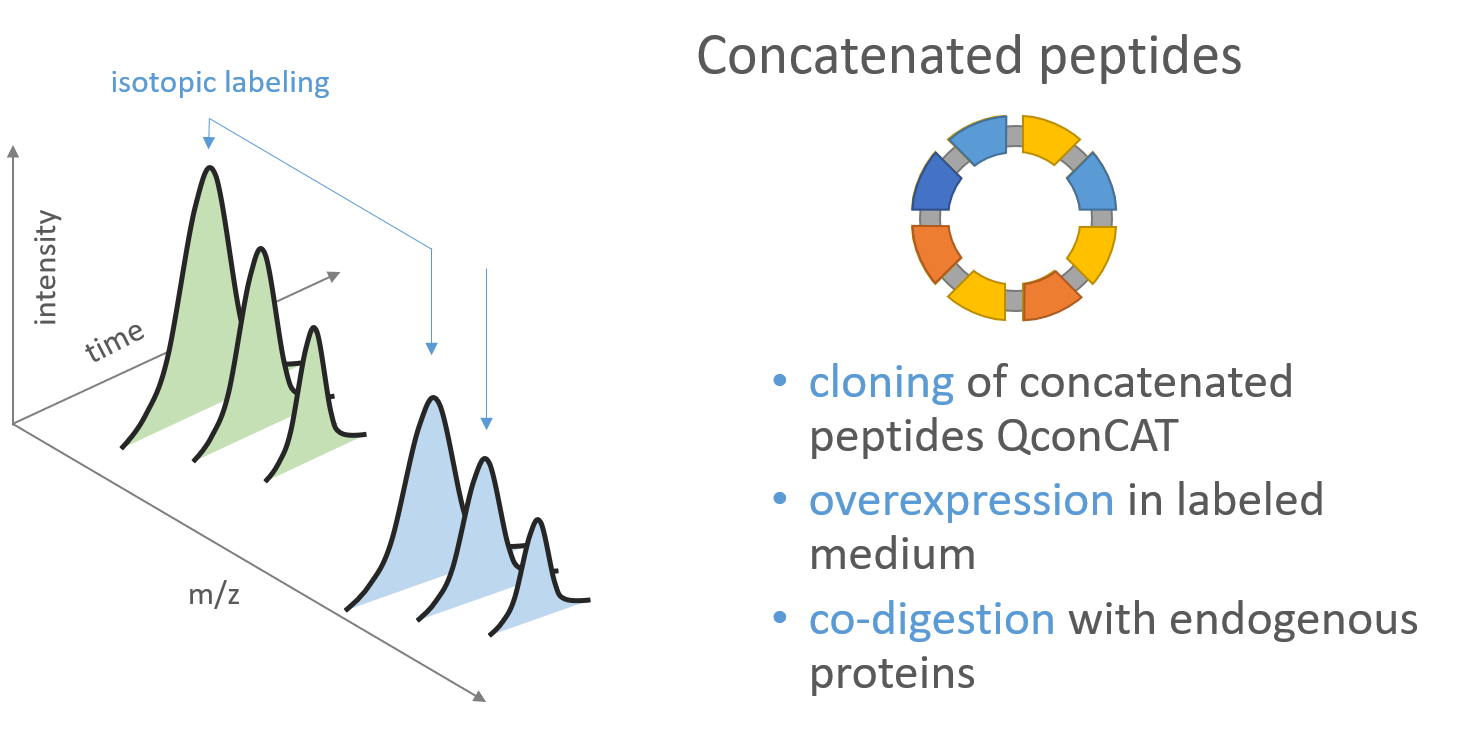
How Do QConCATs work?
Since the design of QConCATs is a research field itself I want to just give a quick insight on how they work in theory. QConCATs are in-silico designed artificial proteins containing several peptide sequences each mapping uniquely to a protein of interest. If the QConCATs is now digested and one assumes a homogenous digestion accross the protein, one can measure the protein concentration of the QConCAT and divide it by the number peptides and thus estimate the amount of a single peptide. If the QConCAT is now labled using 15N (heavy nitrogen) as a nitrogen source, this peptide cocktail can be spiked into samples originating from unlabled nitrogen source and the sample can be analyzed in a single MS run. Since the absolute amount of a single peptide is known, the ratio (e.g. light/heavy) can be used to calculate the absolute protein amount present in the sample.